Image Analysis
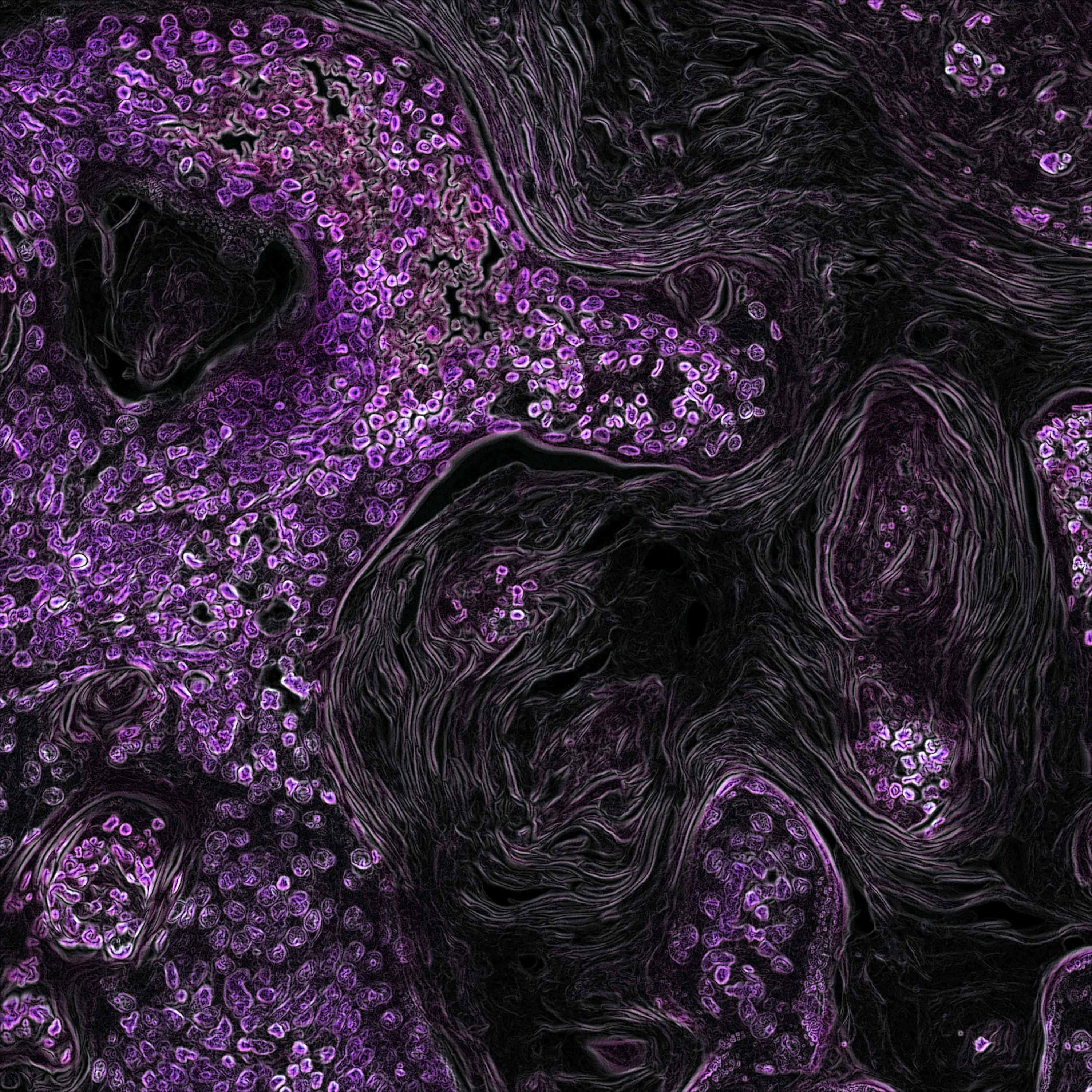
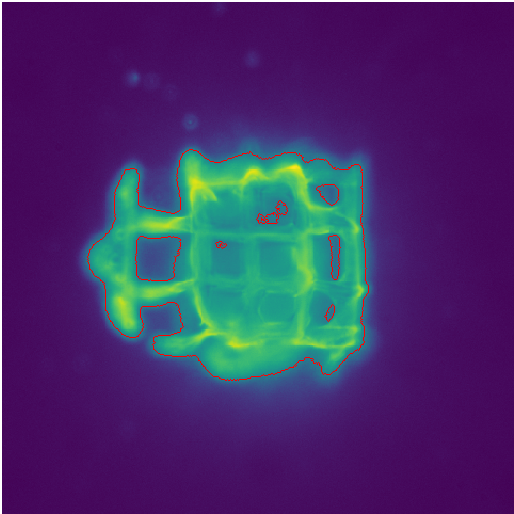
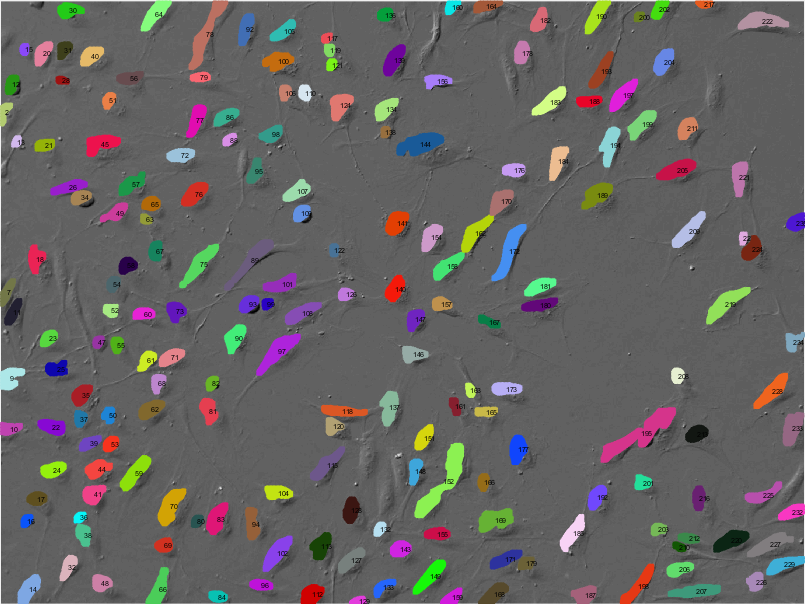
In the Image Analysis research area, we develop and apply methods for computational processing and analysis of digital microscopy images, especially for solving problems in biology and medicine. A wide range of biomedical research projects rely on microscopy data which require specifically designed image analysis workflows to support the identification of complex biological processes. In close collaboration with our partners from research and industry, we develop algorithms and software that analyze microscopy images, for example fluorescence microscopy images of cells or other kinds of organic material. Our focus in algorithm development is mainly on methods of signal processing, machine learning, and computer vision for information extraction and interpretation. Besides filtering and denoising for pre-processing and feature extraction methods, we use machine learning approaches such as deep learning for object detection, segmentation, and object tracking. In addition to workflows and algorithms, we implement image analysis software (e.g. Spotty) for biological research to automatically detect, track, and analyze cells and particles to support fundamental research and drug development. For image analysis, we build on tools and frameworks such as MATLAB, OpenCV, and TensorFlow.
Selected Publications
| 2021 | Jonas Schurr, Christoph Eilenberger, Peter Ertl, Josef Scharinger, and Stephan M. Winkler: | Automated Evaluation of Cell Viability in Microfluidic Spheroid Arrays | Proceedings of the 10th International Workshop on Innovative Simulation for Healthcare, 18th International Multidisciplinary Modeling & Simulation Multiconference 2021 | View Article |
| 2016 | Daniela Borgmann, Sandra Mayr, Helene Polin, Susanne Schaller, Viktoria Dorfer, Christian Gabriel, Stephan Winkler, and Jaroslaw Jacak: | Single Molecule Florescence Microscopy and Machine Learning for Rhesus D Antigen Classification | Scientific Reports 6 | View Article |
