Pre-processing methods¶
IMPI requires as input paired-end reads in *.fastq file format. IMPI allows for processing single files and batches of *.fastq files.
For pre-processing IMPI requires a reference gene sequence (nt) and forward and reverse primers. Primers can contain unique molecular identifiers (UMIs).
Data pre-processing can be invoked by clicking on the Start... button
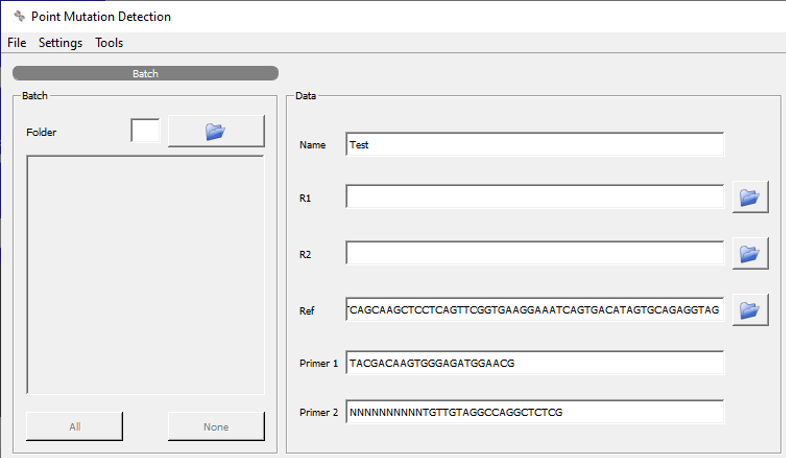
Data input sections for batches of files (left - enter path to folder containing target files) or single file input (right) in paired-end *.fastq file format.
Additionally, the reference gene (nt-sequence or in *.fasta file format) and the primer sequences are mandatory input for successful pre-processing.
UMI nt sequence should be attached to primer sequence using N.¶
Read assignments¶
Overview of the progress of the read counts in IMPI after invoking data pre-processing and application of clusterings.
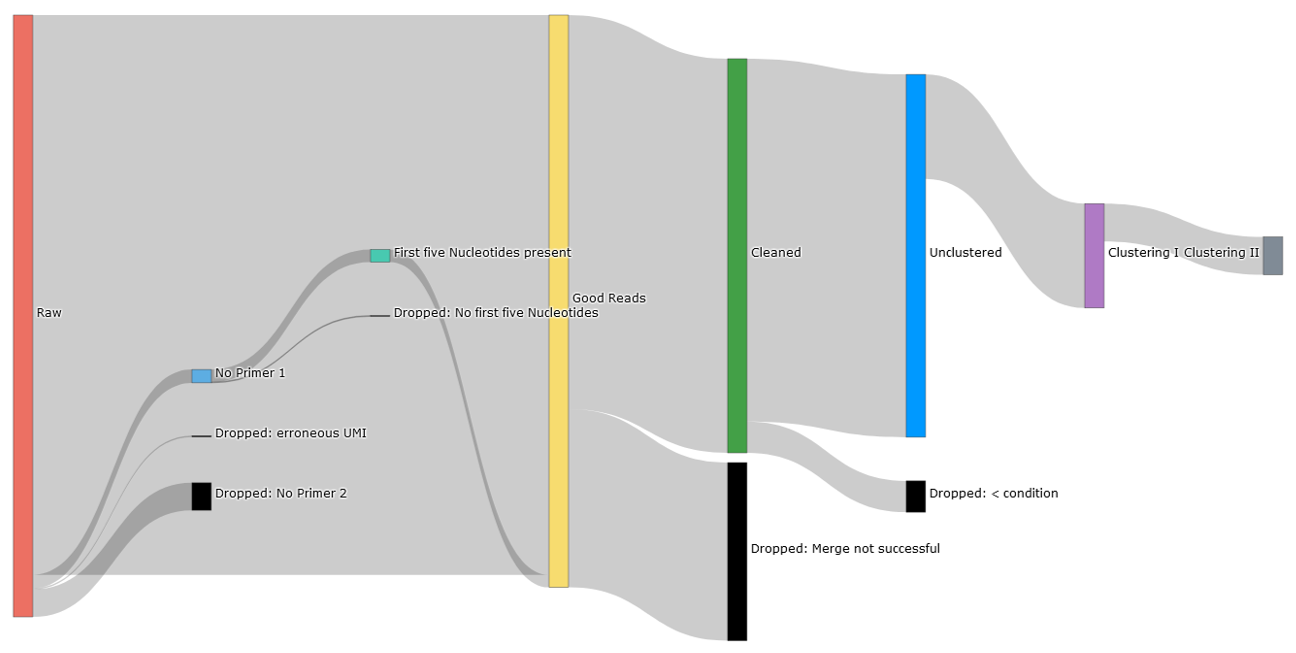
Read assignment within pre-processing and clustering steps - raw read counts (left), read counts after primer und UMI check (yellow), read counts after merge (green), read counts after excluding reads by specific conditions set using the parameter settings (blue), read counts after UMI assembling (violet) and UMI assembling II (gray)¶
PWM information¶
Specific features are extracted and calculated from the SAM output file derived from Bowtie2 mapping and stored in a tab-delimited .csv file. The following features are extracted and used for calculating the position weight matrices later-on in the Allele frequency analysis.
Feature Name |
Description |
|---|---|
ID |
Every read in a FASTQ file has a sequence identifier - this line commonly begins with |
UMI |
Extracted unique molecular identifier sequence which was part of the primer |
UMI_Phred_Quality |
Phred quality scores of the UMI encoded in ASCII characters |
UMI_Avg_Score |
Average Phred quality score of the UMI |
Seq |
Nucleotide sequence of the read with its deletions and insertions which are defined |
Phred_Quality |
Phred quality scores of the whole nucleotide sequence encoded in ASCII characters |
Avg_Score |
Average Phred quality score of the sequence |
Start |
Start position of the sequence within the reference gene sequence |
Length |
Length of the sequence |
Insertions |
Number of insertions |
Deletions |
Number of deletions |
RefIdentical |
Identity of the sequence with the reference gene sequence |
RefMismatches |
Number of mismatches in comparison with the reference gene sequence |
aaSeq |
Translated nucleotide sequence to get the amino acid sequence |
aaRefIdentical |
Identity of the amino acid sequence with the amino acid reference sequence |
aaRefMismatches |
Number of mismatches in comparison with the amino acid reference sequence |