Module: UMI-tools analysis¶
IMPI additionally allows for PWM calculation of reads clustered using the open source tool UMI-tools available here. Learn more about UMI-tools. In the open source UMI-tools software package, there are methods integrated for the identification of errors within the PCR duplicates using UMIs on Linux devices or macOS.
For using the UMI-tools method on Windows there are two ways in IMPI. For both Ubuntu on WSL, also available in Microsoft® Store, has to be installed. Once installed please execute the following commands in the Ubuntu terminal for installing Python
sudo apt-get install python3
and UMI-tools:
pip install umi_tools
Subsequently, IMPI is able to read and convert the output of UMI-tools and generate the desired PWMs.
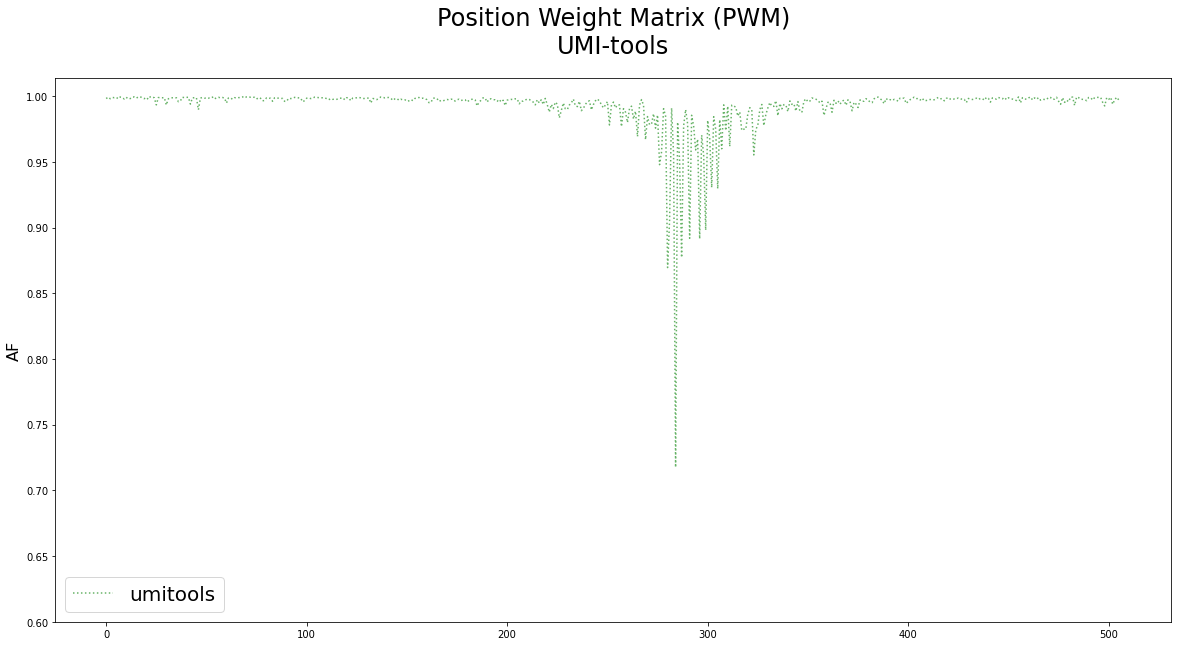
Allele frequencies of the reference nucleotides after PWM generation of reads clustered using UMI-tools.¶
Warning
IMPI has been implemented and tested using Ubuntu on WSL version 2004.2021.222.0. Please make sure you download Ubuntu on WSL in version 2004.2021.222.0 from Canonical Group Limited, otherwise IMPI’s UMI-tools analysis feature might not work properly.
Warning
IMPI has been implemented and tested using UMI-tools version 1.1.1. Please make sure you download UMI-tools in version 1.1.1, otherwise IMPI’s UMI-tools analysis feature might not work properly.
Warning
IMPI has been implemented and tested using Python version 3.7. Please make sure you download Python in version 3.7, otherwise IMPI’s UMI-tools analysis feature might not work properly.