Configuration settings¶
Path settings¶
The path to IMPI’s output files as well as to IMPI’s third party software tools has to be set manually. The pre-processing methods provided in IMPI use the following open-source tools: NGmerge and Bowtie2. You can find more information about third party software and the downloads in the section Third party software.
The path to these third party tools have to be defined in the Settings menu. NGmerge and Bowtie2 commands will be executed automatically within data pre-processing. For mapping, Bowtie2 requires a library containing a reference gene for mapping. Libraries can be build using the bowtie2-build command.
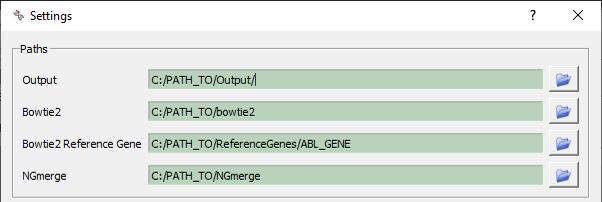
IMPI path settings¶
You can find more information about third party software and the downloads in the section Third party software.
Data specific settings¶
IMPI requires information about the input data. This includes the length of the target sequence and the start position in the reference gene sequence (e.g., c.717). Further IMPI allows the definition of mutation loci of interest (location and mutation name). The allele frequencies of these mutations will be displayed separately in the results view.
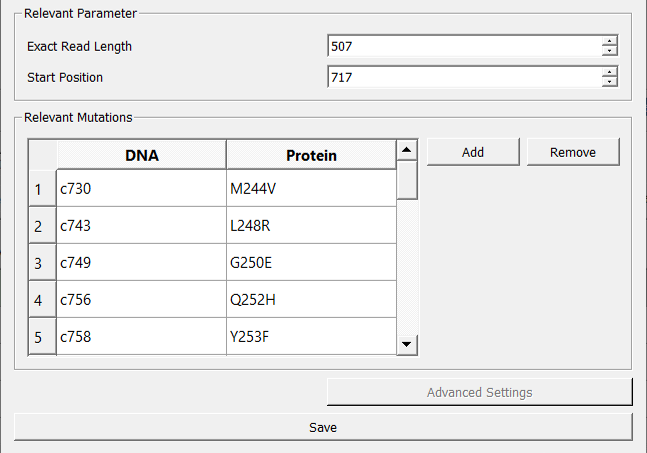
IMPI data specific information¶