Allele frequency analysis¶
Data are cleaned in the pre-processing step of IMPI. Within this step all sample specific information is collected in tab-delimited .csv file format. This file is mandatory for the following analysis and visualisation methods:
All methods provide position weight matrices of the analyzed data. For including or excluding reads by specific conditions defined in the parameter settings section of IMPI can be applied (see Paramter settings).
Paramter settings¶
IMPI allows for custom parameter settings for read selection to generate the position weight matrices (PWMs) containing the allele frequencies of each gene locus.
Parameter Name |
Description |
|---|---|
Min. Quality |
All sequences with a lower average Phred quality score are excluded for |
Min. UMI Quality |
All sequences with a lower average Phred quality score of the UMI are excluded |
Min. Clustersize |
When clustering algorithms are applied, all clusters with a size lower than |
Min. Cut-Off and |
The minimal cut-off value defines the rate of identical nucleotides of the reads |
Max. N |
When a nucleotide at a specific position is not confirmed and is defined as N – |
Threshold Ref Identity |
Only sequences with a reference gene correspondence are taken for PWM |
Max. Mismatches, |
Definition of the maximal number of mismatches, insertions or deletions |
Omit from UMI |
Since the first few nucleotides within a read show rather low quality IMPI allows |
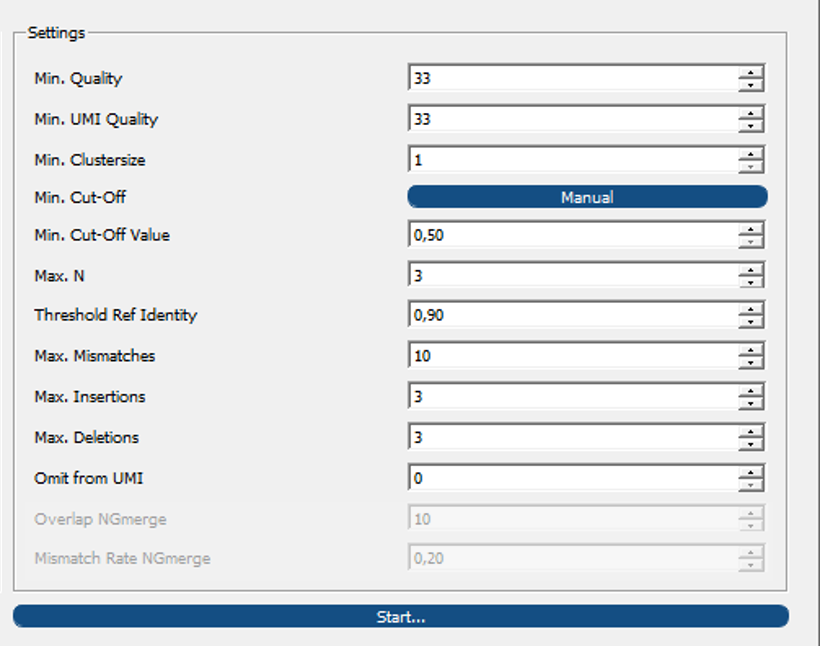
Parameter settings section of IMPI for custom parameter configurations for PWM generation and read sorting out.¶
Raw read analysis¶
IMPI allows for automated processing and evaluation of NGS data, focusing on the detection and identification of point mutations and provides information about allele frequencies (AF). After providing the custom settings described in the previous section Paramter settings, IMPI calculates allele frequencies at each position using the reads which fulfill all conditions set and displays the resuls in a PWM.
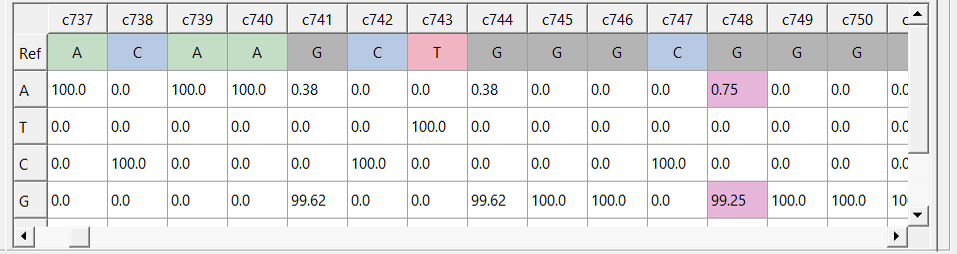
Position weight matrix (PWM) showing the allele frequencies at specific loci and the reference sequence. Values exceeding a custom set min_val (here: 0.5% AF) are colored in violet.¶
Clustering I and II¶
IMPI integrates two clustering steps for straightforward clustering but is further able to process files derived from clustering using UMI-tools. In a first step, the straightforward clustering (Clustering I) groups the raw sequences by their UMI and builds consensus sequences. In the second clustering step, all clusters containing fewer reads than the previously defined minimal cluster size undergo an additional re-clustering (Clustering II). UMIs that are up to 90% identical are clustered together.
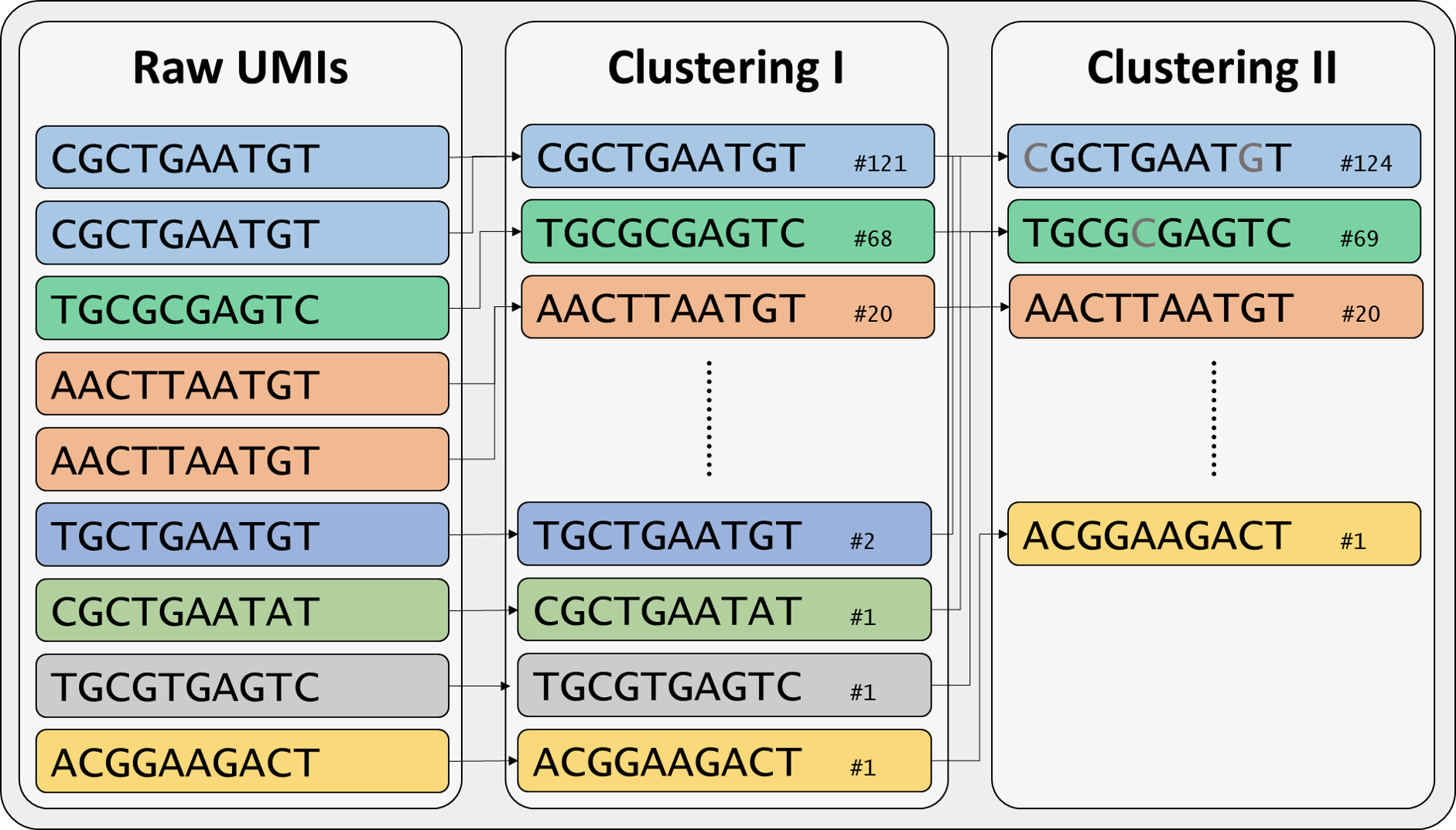
Clustering method integrated in IMPI using UMIs¶
Clinically relevant mutations¶
Since IMPIs’ functionality has been tested using chronic myeloid leukemia (CML) data, IMPI provides the possibility for previously define clinically relevant mutations (see section Data specific settings). IMPI automatically collects and displays a PWM containing only clinically relevant mutation data.
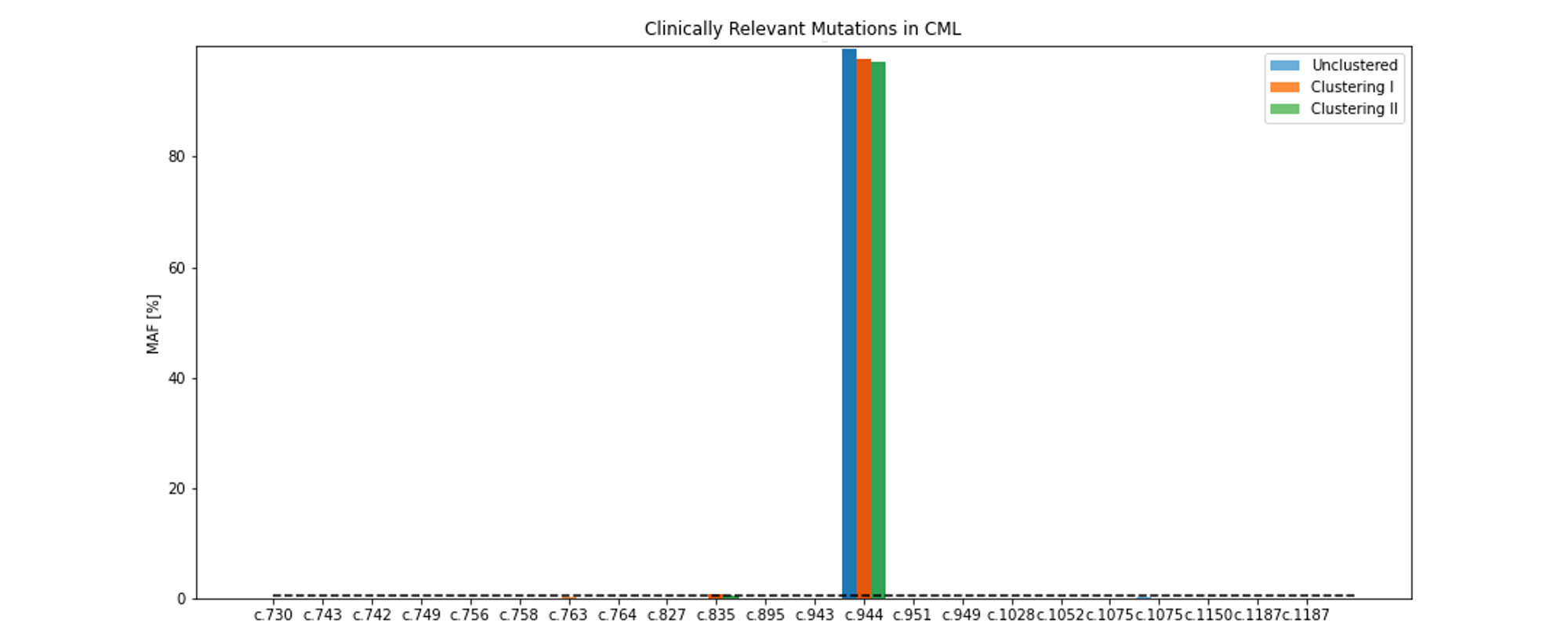
Allele frequencies of minor allele frequencies (MAFs) of clinically relevant mutations (resistance mutations) in CML patients displayed as barchart.¶