Preprocessing methods
The preprocessing methods provided in IMDA use the following open software tools: MIGEC, MiXCR and Bowtie2. You can find more information about third party software and the downloads in the section Third party software.
do_MIGEC and do_MiXCR require *.fastq or *.fastq.gz files as input. Furthermore, MIGEC requires a tab-delimited barcodes.txt file. MiXCR allows the use of other libraries. IMDA has been tested using the imgt.201802-5.sv2 library.
Bowtie2 requires *.fastq or *.fastq.gz files and a library. Libraries can be build using the bowtie2-build command. Further we provide a prepared library included in Download with PhiX genome and various TCR sequences.
For the FACS error correction module a tab-delimited pairs.txt file is required.
Format Description
mixcr_library = “–library D:/Forschungsgruppe/KidTrans/MiXCR/imgt.201802-5/imgt.201802-5.sv2”
barcodes.txt and pairs.txt have to be provided in case if MIGEC or the FACS error correction module is used, respectively.
barcodes.txt
| #SampleID | Master barcode sequence | Slave barcode sequence | FASTQ#1 | FASTQ#2 |
|---|---|---|---|---|
| 1_A_nS_r1 | NNNNNNtCAGATtNNNNNNtcttgggg | idx1_R1_001.fastq.gz | idx1_R2_001.fastq.gz | |
| 1_A_nS_r2 | NNNNNNtCAGATtNNNNNNtcttgggg | idx2_R1_001.fastq.gz | idx2_R2_001.fastq.gz | |
| 1_B_nS_r1 | … | … | … |
pairs.txt
| #SampleID1 | SampleID2 |
|---|---|
| 1_A_nS_r1 | 1_A_nS_r2 |
| 1_B_nS_r1 | 1_B_nS_r2 |
| … | … |
Read Assignment Visualisation
For an immedeiate overview of the progress of the read counts IMDA provides a sankey diagram (Figure 1) showing the raw read counts, the read counts after read assignment by barcode and the read counts after the UMI assembling within MIGEC.
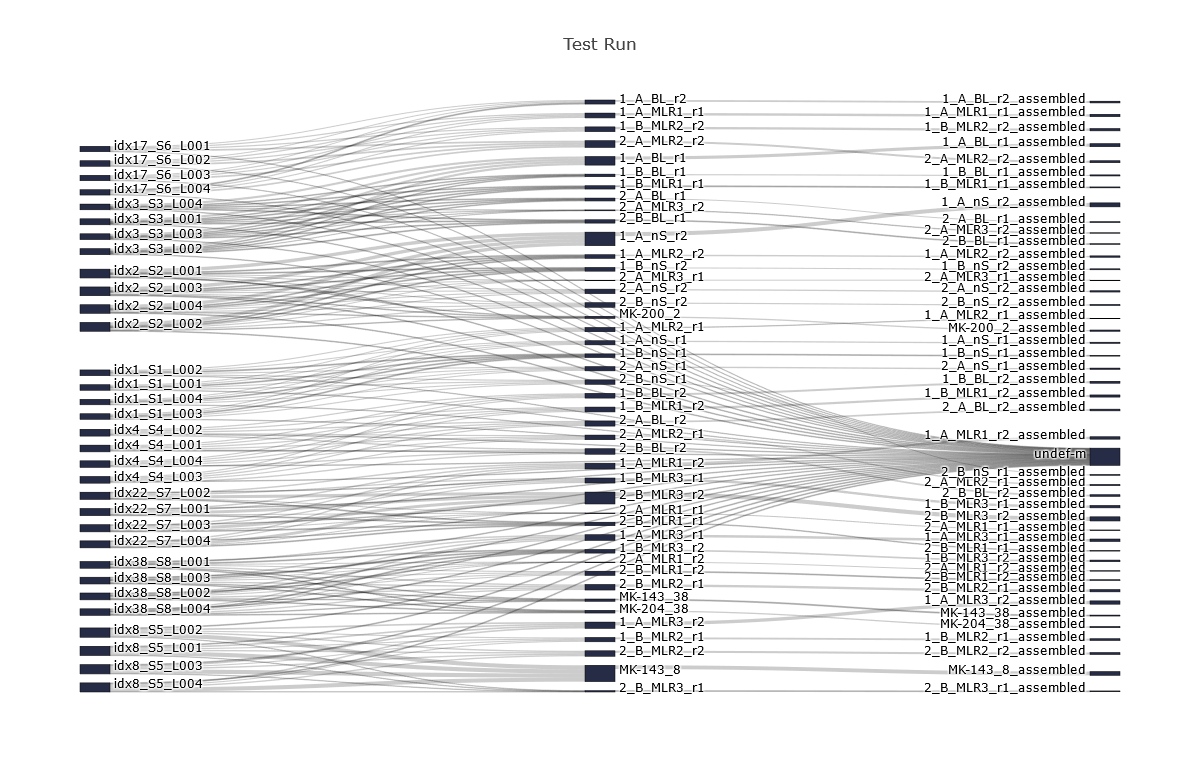
Figure 1: Read assignment within MIGEC - raw read counts (left), read counts after read assignment by barcode (middle), read counts after UMI assembling (right)