Module: Sample Contamination Analysis
IMDA Prep includes besides the FACS error correction module, a cross-sample contamination analysis module. This analysis is based on UMIs. If MIGEC is used for data preprocessing, this module can be used immediately.
If MIGEC is not used for preprocessing, the reads within the FASTQ files require the following ID format: @ReadID RX UMI:NNNNNNNNNNN:Y
@MIG.132988 R1 UMI:TAGTCGACGACG:2
ACAGTGACCCTGATCTGGTAAAGCTCCCATCCTGCCCTGACTCTGTCATGGGCACCAGGCTCCTCTGC
+
IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII
The number of shared clonotypes is stored within the summary files and visualised using Venn diagrams (Figures 9A and 9B).
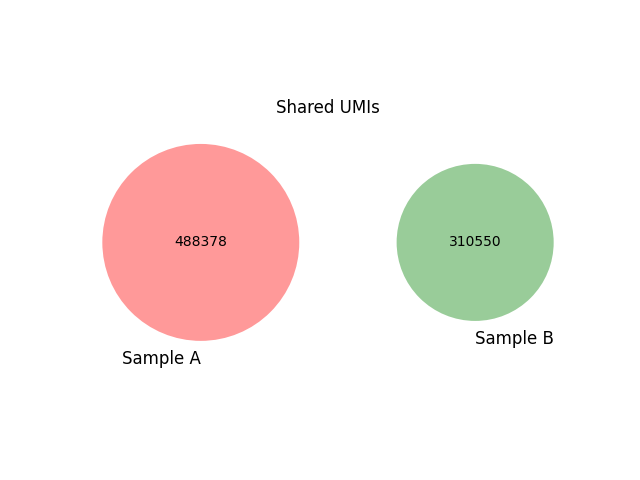
Figure 9A: Cross-sample contamination analysis |
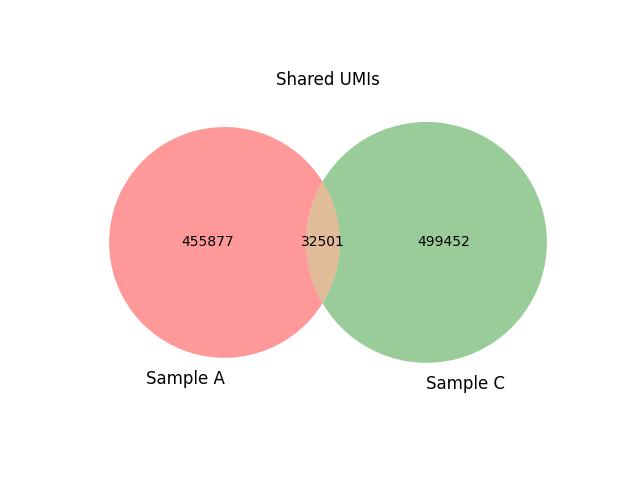
Figure 9B: Cross-sample contamination analysis |